
|
BATS - The Models |




|
Bayesian Analysis for Time Series Microarray Experiments |


|
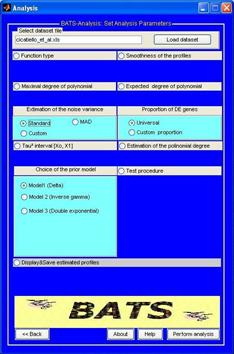
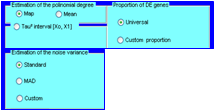
Though these sub-windows an user can choose one of the three prior models implemented in BATS and can choose how to estimate the hyper-parameters. |

|
We distinguish 3 Models Model 1)
Model 2)
Model 3)
|
|
the marginal distribution of the noise Student T |
|
Home page |
|
About Us |
|
Download BATS |
|
Description |
|
Tutorial |
|
Related Papers |
|
FAQ |
|
Acknowledgments |
|
Contact us |
|
A user friendly software for Bayesian Analysis of Time Series Microarray Experiments. |

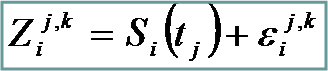
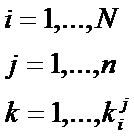
|
Number of time-points |
|
Number of replicates |
|
Number of genes
|
|
Observed data |
|
Gene “true” functional profile
|
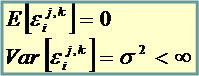
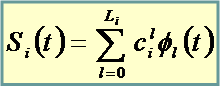
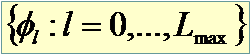
|
Noise. i.i.d. |


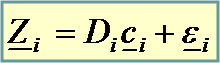
|
We assume that genes are conditionally independent
|
|
And we place a prior on unknown parameters
|

|
Poisson truncated at
|
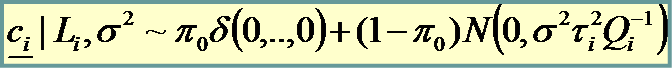
|
Gene “non affected” by the treatment |
|
Gene “affected” by the treatment
|

|
Gene’s specific variance (to be estimated from the data) |
|
Prior probability of not being affected by the treatment (to be estimated from the data) |

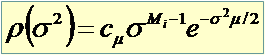
|
the marginal distribution of the noise is Gaussian |
|
the marginal distribution of the noise Double-exponential
|
|
Noise model in BATS |
|
Data model in BATS |
|
Model + observed data
|
|
Prior Information
|


|
Posterior Distribution
|

|
For 1)-3) it is analitically known. Moreover hyper-parameters can be estimated from the data
|
|
C. Angelini, D. De Canditiis, M. Mutarelli, M. Pensky. A Bayesian Approach to Estimation and Testing in Time-course Microarray Experiments, Statistical Applications in Genetics and Molecular Biology: vol 6 : Iss. 1, Article 24, (2007). |
|
For a detailed description of the statistical method implemented in BATS the users are referred to the following paper |

|
Global parameters
Can be either estimated from the data or chosen by the users Gene specific parameters can be estimated from the data as in the empirical Bayes approach
|
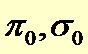









![Fumetto 4: The expansion is in an orthogonal system on [0,T]. Legendre and Fourier bases are currently available. Suggested value is n/2 where n is the number of different time points](image3082.png)


|
For models 1)-3) the posterior distribition can be analitically evaluated and it is possible to test the statistical hypothesis |
|
|
|
|
|
VS |
|
Statistical decision (both selection and ranking) can be taken by looking at the Bayes Factors of each gene |
