|
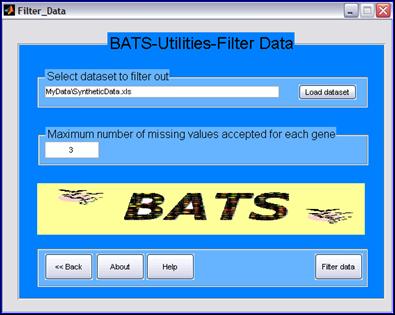
Before starting the analysis, BATS will check for missing points in data and will output an warning message if too many missing values are included in the dataset In the Utilities menu of BATS the function Filter Data can be used to remove genes with an excessive number of missing points and then filtered data can be used at next stage of the Analysis. |
|
After the choice of the maximum number of missing points to be retained for each gene, by clicking the Filter data button the software will create a new file with the string original_file_name_filter_m.xls attached to the end of the input file name, containing the filtered data (where m denotes the maximum number of missing data allowed in each gene record). |



|
Bayesian Analysis for Time Series Microarray Experiments |

|
A user friendly software for Bayesian Analysis of Time Series Microarray Experiments. |